An Introduction to Linkage Analysis
Introduction
Linkage analysis is a method that is used in establishing the carrier status of female 'at-risk' carriers and for prenatal diagnosis. In many/most cases, linkage analysis has been replaced by mutational analysis but in a small number of families in whom the mutation cannot be identified, linkage analysis remains the only method for the genetic diagnosis of carriers.
Linkage: Two genetic loci are said to be in linkage if the alleles at these loci segregate together more often that would be expected by chance – that is the two loci are so close together on the same chromosome that the chances of them separating by a crossover event (recombination) during Meiosis is small. The probability that any two alleles at two randomly selected loci with be inherited together is 0.5. If two loci are closely linked then the chances of a crossover or recombination event occurring is <0.5. The chances of recombination taking places is linked to the distance between any two loci. The recombination fraction [θ] is a measure of the genetic distance between two loci. The distance between two loci is measured in centimorgans and 1 centimorgan is defined as the genetic distance between two loci with a recombination frequency of 1%. Although the centimorgan is not a measure of physical distance, it typically equates to a physical distance of one million base pairs. So two loci close to the F8 gene with a 5% probability of recombination would be 5 centimorgans apart i.e. approximately 5 million base pairs.
The aim of linkage analysis is to identify a marker that co-segregates with the gene of interest and so can be used to track the gene within a family without actually knowing the mutation. By definition this marker must co-segregate with the gene of interest and so be present in affected family members but absent in unaffected family members. In the era before rapid sequence analysis, linkage analysis was the principal method for establishing the carrier status of 'at-risk' females within a family and for pre-natal diagnosis.
Whilst we usually think of linkage analysis using DNA markers, markers such as proteins can be also be used. Such a case is the gene for Glucose-6 Phosphate Dehydrogenase [G6PD] which maps to the long arm of the X-chromosome at Xq28 close to the gene for Factor VIII [F8]. Close linkage between the loci for G6PD and F8 has allowed prenatal diagnosis of Haemophilia A in women who are heterozygous for the two electrophoretic variants (A and B) of G6PD.
The pedigree below illustrates the theoretical use of G6PD variants (A and B) for carrier detection in a family with severe haemophilia A.
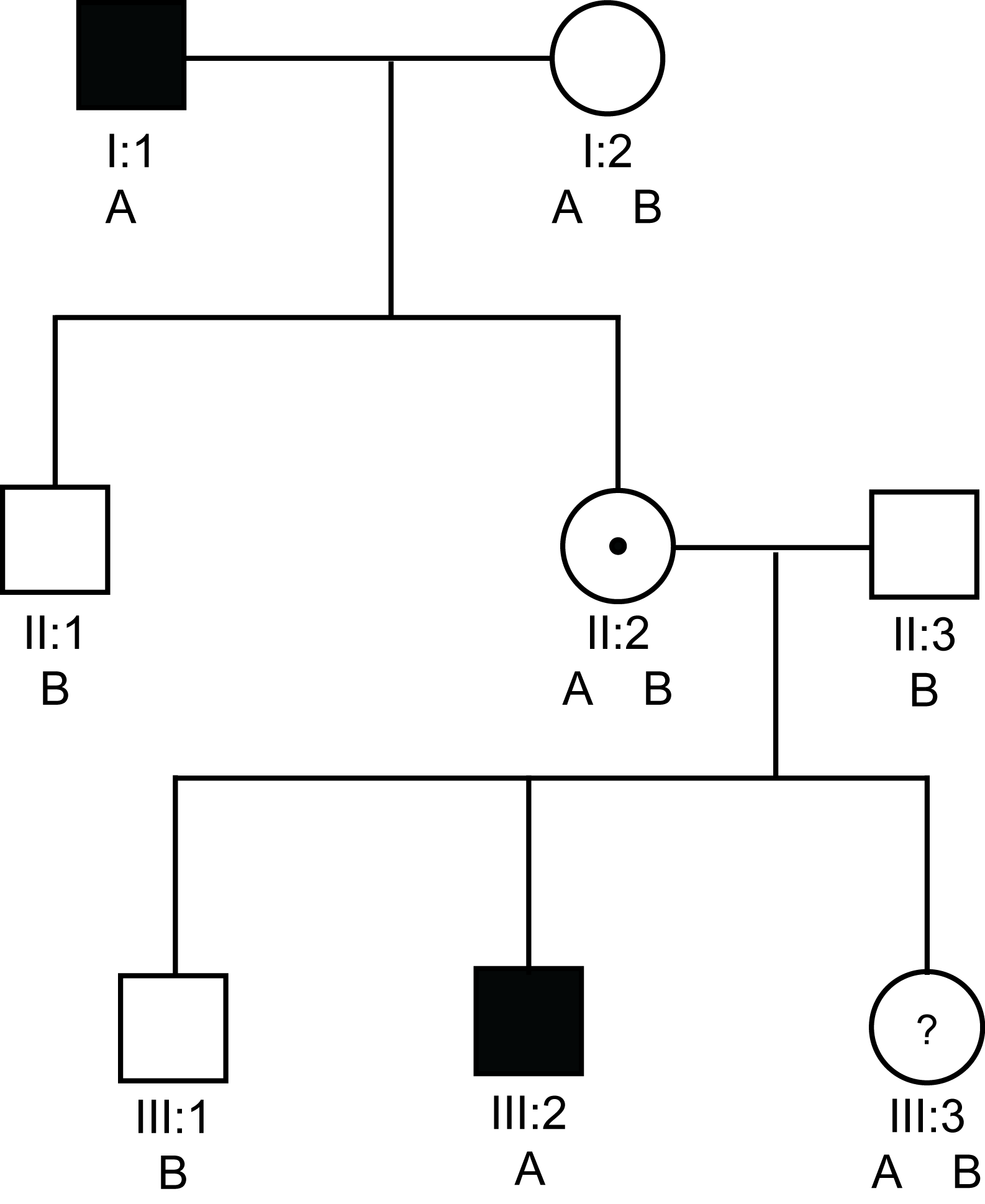
Solid square = a male with Haemophilia
Open square = a male without Haemophilia
Open circle = non-carrier female
Circle with a dot in the centre = a female carrier of an X-linked disorder
Circle with a ? in the centre = a possible female carrier of an X-linked disorder
A & B denote the two possible eletrophoretic variants of G6PD
In this pedigree, I:1 and III:2 have severe haemophilia A [VIII:C<1 IU/dL). II:2 must be an obligate carrier and III:3 wishes to know if she is a carrier or not. From the pedigree there is a 1/2 chance that she is or is not. Analysis shows that they both have the A variant of G6PD. In contrast, the unaffected males in this pedigree have the B variant. So in this family we can use the A variant of the G6PD protein to track the abnormal F8 gene.
If we use the G6PD electrophoretic variants [remember the gene for G6PD is located on the X-chromosome at Xq28 close to the F8 gene which also maps to Xq28] - then III:3 has inherited the B allele from her father and the A allele which tracks with the abnormal F8 gene from her mother and she is, therefore, likely to be a carrier.
Bayesian risk analysis would allow us to make more confident predictions as to her carrier status but to undertake this we would need to know the frequency of recombination occurring between the F8 gene and the G6PD gene. Furthermore, measurement of the FVIII:C and VWF:Ag ratio would allow us to derive a VIII:C/VWF:Ag ratio and this may allow us to more accurately predict the carrier status for III:3.
There are, of course, serious limitations to this method of linkage analysis and in particular the risks of a recombination event occurring with each generation and as a result incorrectly assigning the carrier status of ‘at-risk’ female. In addition, it relies upon the identification of women who are heterozygous for variants of G6PD. This is found in approximately 40% of black females in the USA but uncommon in other ethnic groups.
Polymorphic DNA Markers
We have seen how we can use protein variants to track a gene within a family but more commonly we use DNA markers.
The aim of linkage analysis is to identify a DNA marker that co-segregates with the gene of interest and so can be used to track the gene within a family without actually knowing the mutation. The markers that we now commonly use to track a gene within a family are known as polymorphic markers or polymorphisms. There are various types of polymorphisms
A. Single Nucleotide Polymorphisms [SNPs]
B. Short Tandem Repeat [STRs] or Variable Number Tandem Repeats [VNTRs]
A. Single Nucleotide Polymorphisms [SNPs]: Are single nucleotide changes that usually, although not always result in no change to the amino acid sequence of the protein of interest. Polymorphisms are located throughout out the human genome and can be found both within a gene (so-called intragenic polymorphisms - usually within the introns of a gene or in the immediate 5' and 3' untranslated regions [upstream of downstream of the coding sequence of a gene]) or closely linked to a gene (so-called extragenic markers).
The further a marker is from the gene of interest, the greater the chance that recombination will occur during meiosis.
Historically, SNPs were often designated by the restriction endonuclease or enzyme which was used to digest the DNA prior to agarose gel electrophoresis and Southern Blotting. For example within the F8 gene the enzyme Bcl I identifies an intragenic polymorphism located within intron 18 and which cuts the DNA into two sequences and which gives rise of 2 fragments of 0.8kb and 1.1kb when the digested DNA fragments are resolved on agarose gel, blotted on nylon membranes and then probed with a labelled DNA fragment that binds to the DNA sequences of interest. Similarly the enzyme Bgl I identifies a SNP located within intron 25 of the F8 gene and cuts the DNA into two sequences of 5kb and 20kb whilst the enzyme Bgl II identifies a SNP located close to but not part of the F8 gene and gives rise to two fragments of 5.8kb and 2.8kb.
The common feature is that when digested with a restriction endonuclease e.g. Bgl II, Bcl I, Bgl I etc, DNA products are generated which are of different lengths. The polymorphisms giving rise to these differing fragments are known as Restriction Fragment Length Polymorphisms or RFLPs. Click HERE for more information on RFLPs.
Southern blotting is rarely performed today and most SNPs are detected by PCR with either sequence analysis or resolution of the DNA fragments on gel electrophoresis. For a review on Southern Blotting - click HERE.
B. Short Tandem Repeat [STRs]: STRs are very useful DNA markers as they are highly polymorphic and inherited in a strictly Mendelian fashion.
Areas of repetitive DNA occur throughout the genome where the repeating unit is very small, usually 1-6 nucleotides. These are generally polymorphic within a population and can be used for bone marrow transplant engraftment, forensics, identity testing, paternity testing etc. Common STRs include dinucleotide repeat sequences e.g. [CA] in which the repeated sequence occurs multiple times a e.g. [CACACACACA] and are notated, therefore, as [CA]. Other STRs include trinucleotide repeats e.g. [ATT]n or tetranucleotide repeats e.g. [GATA]n. STRs are widely used in genetic linkage studies and the reason for this lies in the greater chance that a particular individual may be heterozygous for a particular marker. Although the number of repeat sequences can change - this happens only every 100 generations or so.
Click STR or VNTR for information.
The pedigrees below illustrates linkage analysis using two hypothetical polymorphisms:
Pedigree 1a: Linkage analysis using a SNP.
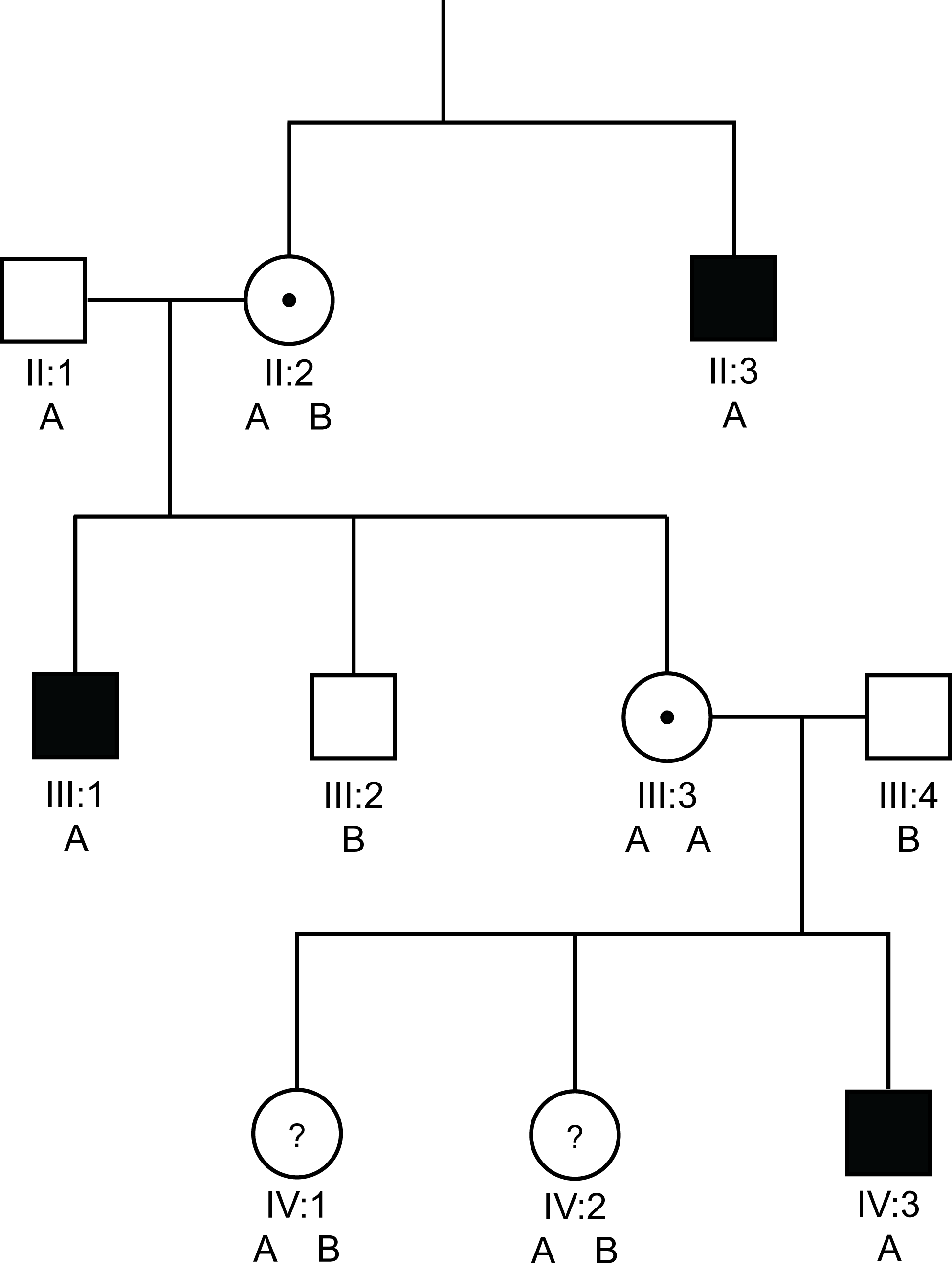
Solid square = a male with Haemophilia
Open square = a male without Haemophilia
Open circle = non-carrier female
Circle with a dot in the centre = a female carrier of an X-linked disorder
Circle with a ? in the centre = a possible female carrier of an X-linked disorder
A & B denote the two possible alleles the SNP
In the pedigree above we have undertaken linkage analysis using a SNP that has two possible alleles - A or B. As we are looking, in this case at the F8 gene - males are hemizygous that is they have only a single X-chromosome and so can can have only a single SNP [A or B] whilst females possess 2 X chromosomes and so can have three possible combinations - homozygous AA, homozygous BB or heterozygous AB.
In this pedigree with severe haemophilia A, we can see that the abnormal F8 gene is marked by the A allele of our SNP. The males in the family with Haemophilia A, shown by the solid squares, all have the 'A' allele of the SNP whereas the male without Haemophilia A, male III:2 has the 'B' allele.
II:2 has to have both the A and B alleles i.e. she is heterozygous for the polymorphisms - so that she can have two sons with differing genotypes.
III:3 must inherit the A allele from her father [he has only a single X chromosome] and she has inherited the A allele from her obligate carrier mother II:2 - so III:3 must be a carrier and indeed this is confirmed by the finding that she has a son IV:3 with severe Haemophilia A. However - we could not use this polymorphism for pre-natal diagnosis in III:3 as she is homozygous AA and so we would be unable to establish which of the two A alleles tracked with abnormal F8 gene.
In the cases of IV:1 and IV:2 - both must inherit a B allele from their father but again it is not clear which A allele has been inherited from their mother [III:3] i.e. the one that tracks with abnormal F8 gene or the other. This SNP cannot, therefore, be used to establish the carrier status of IV:1 or IV:2.
Pedigree 1b: Linkage analysing using a VNTR
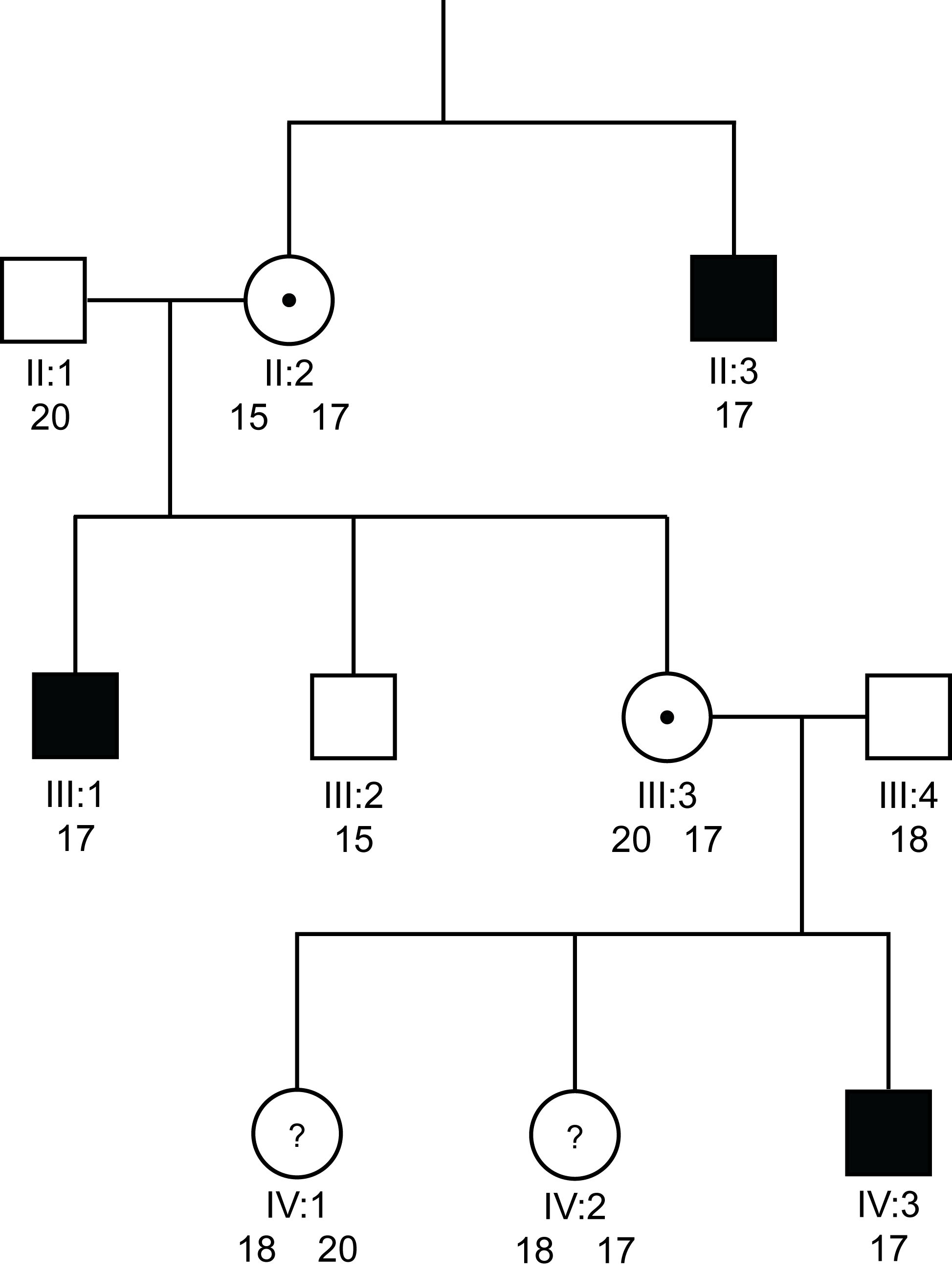 >
>
Solid square = a male with Haemophilia
Open square = a male without Haemophilia
Open circle = non-carrier female
Circle with a dot in the centre = a female carrier of an X-linked disorder
Circle with a ? in the centre = a possible female carrier of an X-linked disorder
Numbers denote the possible alleles for the VNTR
In this pedigree which is the same as Pedigree 1a, we have used an VNTR - in this case the repeat sequence - [GT]n located within intron 1 of the F8 gene. Again males can only have a single copy of this sequence but females can have various combinations depending upon the number of repeat sequences.
We can see that the abnormal F8 gene is marked by the 17 repeat sequence of the [GT]n VNTR [i.e. there are 17 GT repeats with intron 1 of the F8 gene in III:1, IV:3 and II:3] whereas the male III:2 without Haemophilia A, has the 15 repeat allele. II:2 has to have both the 15 and 17 alleles so that she can have two sons with differing genotypes.
III:3 must inherit the 20 repeat allele from her father [he has only a single X chromosome] and she has inherited the 17 repeat allele from her obligate carrier mother II:2 - so III:3 must be a carrier and indeed this is confirmed by the finding that she has a son IV:3 with severe Haemophilia A. However - we can now use this polymorphism for pre-natal diagnosis in III:3 as she is heterozygous 15/17 and so we know that the 17 repeat allele tracks with the abnormal F8 gene.
In the cases of IV:1 and IV:2 - both must inherit the 18 repeat allele from their father but now we can see that IV:1 has inherited the 18 repeat allele from her mother and so is not a carrier of severe Haemophilia A, whereas IV:2 has inherited the 17 repeat allele and so is a carrier. Furthermore, we can use this [GT]n repeat for pre-natal diagnosis in IV:2.
This pedigree highlights the value of VNTRs in both carrier detection and pre-natal diagnosis. As a result of the variation in copy numbers between individuals when we use VNTRs, there is a greater chance that a female will be heterozygous for a particular marker.
The following illustrations show a variety of SNPs and VNTRs.
1. Electropherogram showing an [ATT]
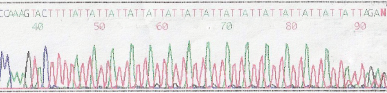
The sequencing gel below again shows the Antithrombin [ATT]n repeat sequence but instead of displaying an electropherogram - the bases are displayed as bands on an autoradiograph.
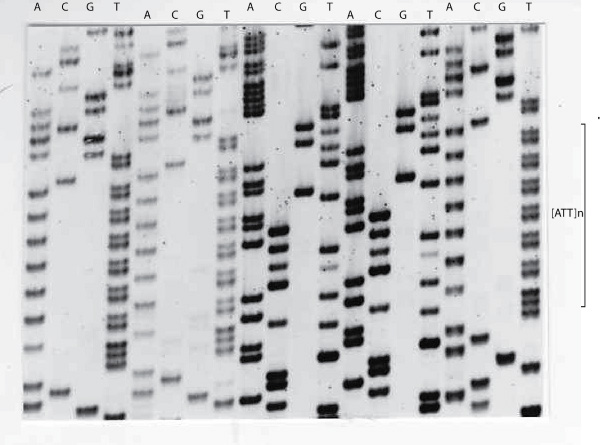
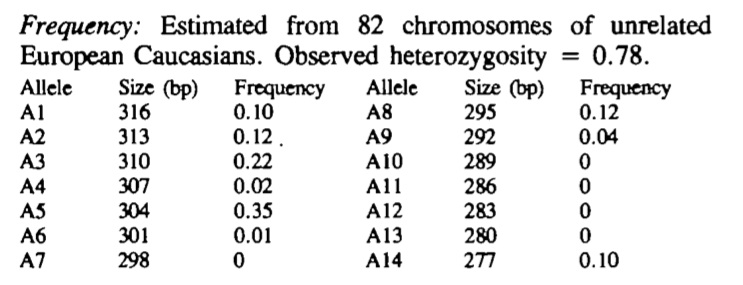
To establish the frequency of the various alleles with this Antithrombin [SERPINC1] gene [ATT]n repeat, primers were designed to amplify the repeat and a series of DNA samples were amplified and the products run on agarose gel. The allelic frequencies are summarised in the table below. In this study 75% of individuals were heterozygous.
What Test Next
In many families, mutational analysis has replaced linkage analysis. However, the results of any genetic test must take into account both pedigree and phenotype data. In approximately 3% of cases of Haemophilia A, a mutation cannot be found and so linkage analysis remains of value in establishing the carrier status of 'at risk' females.
