Factor V Leiden [F5 Arg506Gln]
Introduction
Resistance to Activated Protein C [APCr] was first assessed used a modified APTT-based assay [see APCr assays]. Subsequent studies have shown that in the majority of cases of APCr, it arises from a G→A missense mutation at nucleotide 1691 in the F5 gene leading to an Arg506Gln mutation in which the Arginine residue at position 506 in Factor Va is replaced by a Glutamine and this abolishes an inactivation cleavage site for Activated Protein C [APC] .
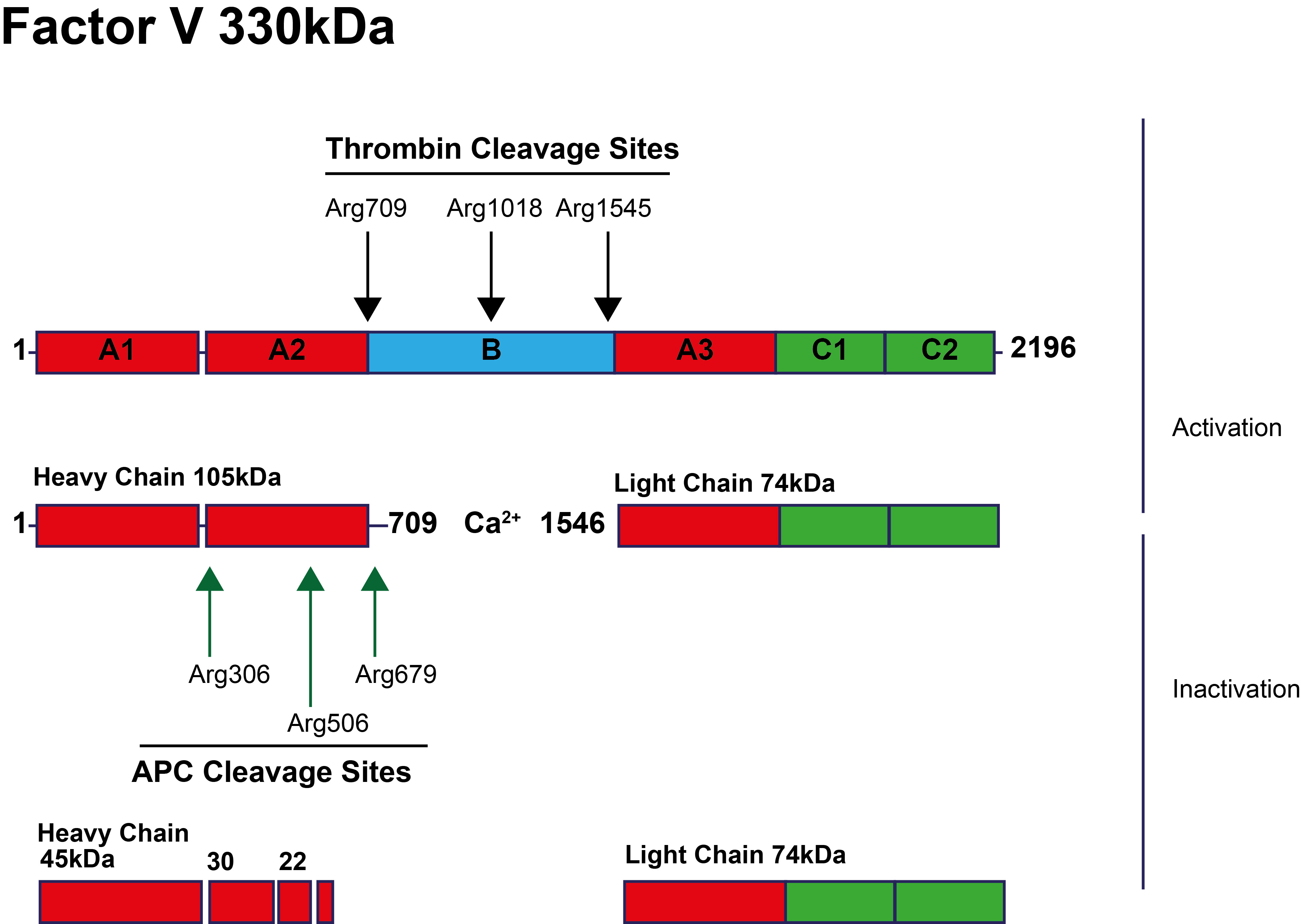
Factor V is activated to Factor Va by the cleavage of specific residues by Thrombin - see illustration below. This gives rise to a Calcium-dependent heterodimer consisting of a Heavy chain and a Light chain. APC-mediated cleavage and inactivation of FVa occurs at specific residues including
Arg506, 306 and 679 and Lys994. The first cleavage at Arg506, causes
partial inactivation of FVa and is essential for optimal exposure
of the subsequent cleavage positions. Subsequent cleavage at
the other cleavage sites, most importantly Arg306, results in
the complete loss of FVa cofactor activity in the Prothrombinase
complex.
In the diagram below the cleavage sites for Thrombin [IIa] and APC are shown:
Principles & Method
A number of methods exist for the detection of the FV Leiden mutation. The majority of methods depend upon PCR to amplify genomic DNA followed by a method to identify the presence or absence of the missense mutation e.g. restriction enzyme digestion and mapping.
A wide variety of methods are in use [see References for additional information]:
- Multiplexed techniques e.g. for the FVL and Prothrombin G20210A mutations
- Direct sequencing, either single pass or bidirectional
- Spectrophotometric evaluation of binding to allele-specific oligonucleotides in microtitre plates
- Enzyme immunoassay for the PCR product
- Luciferase-linked DNA polymerase-mediated depolymerisation
- PCR-independent oligonucleotide-generated fluorescence
- Direct fluorogenic probe-based PCR assay
- Real-Time PCR
- Nanochip micro electronic arrays
Interpretation
Most techniques can rapidly distinguish heterozygous individuals from those who are homozygous for a mutant allele or wild type [i.e. normal]. It is important to remember that the majority of tests only screen for the presence or absence of the Arg506Gln mutation and although an abnormal APC ratio may be obtained in a screening test for APC resistance, in the rare cases in which this is due to a mutation in an APC inactivation cleavage site other than at Arg506, this/these will not be detected unless specific approaches to identify these are undertaken.
Similarly the Factor V HR2 haplotype [His1299Arg found in combination with Met385Thr] has been demonstrated to be associated with reduced Factor V levels and interact synergistically with the Arg506Gln mutation in individuals who are heterozygous for the Arg506Gln mutation.
F5 mutations associated with APC-R but which are not due to the FVL Arg506Gln mutation have also been reported:
| Protein | Mutation | Location |
| Factor V Leiden | Arg506Gln | Exon 10 |
| Factor V Cambridge | Arg306Thr | Exon 7 |
| Factor V Hong Kong | Arg306Gly | Exon 7 |
| Factor V Nara | Trp1920Arg | Exon 20 |
| Factor V Liverpool | Ile359Thr | Exon 8 |
| Factor V Bonn | Ala540Val | Exon 10 |
Heterozygous carriers of the FV Leiden mutation, or other mutations, who have, in addition, a null mutation within the other F5 gene leading to non-expression, results in a pseudohomozygous APC-R phenotype.
Reference Ranges
There are no reference ranges for a genetic test - the mutation is either present in a heterozygous or homozygous form [and rarely a pseudohomozygous form] or it is not.
